The biological detection platform is designed to conduct molecular genetic analysis of complex human diseases, decipher the mechanisms underlying their occurrence and development, so as to improve the diagnosis, treatment and prevention of diseases. It aims to establish a series of advanced population-based biotechnology platforms for genotyping, phenotyping, and integrated omics, and to develop a range of sophisticated biotechnology and clinical detection tools. By utilizing methods such as whole-genome association analysis and other methods based on bioinformatics to identify and verify biomarkers of disease susceptibility, early screening, diagnosis, treatment, therapeutic reactions, disease progression, recurrence, surgical complications, survival time and quality of life and other disease course.
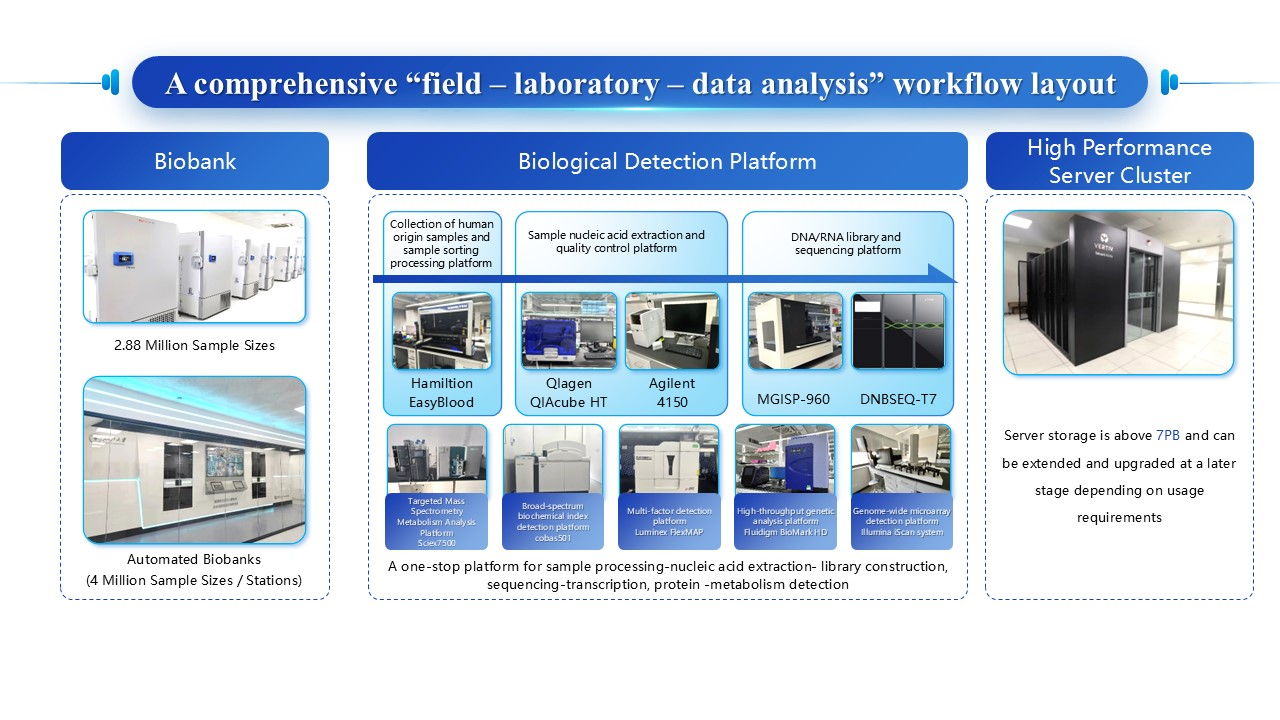
We have built a one-stop platform for sample processing-nucleic acid extraction- library construction, sequencing-transcription factors, protein, and metabolite detection. This platform is equipped with high-throughput detection systems and automation equipment such as the Hamilton EasyBlood automated sample dispensing workstation, QIAGEN automated nucleic acid extractor, fully automated microfluidic analysis system, Illumina iScan gene chip scanning system, DNBSEQ-T7 ultra-high-throughput gene sequencer, MGISEQ-2000 gene sequencer, BioMark HD high-throughput gene analysis system, Luminex liquid suspension chip system, and targeted mass spectrometry metabolic analysis system. Additionally, we possess basic equipment including PCR systems, cell counters, spectrophotometers, high-speed refrigerated centrifuges, thermostatic mixers, and gel imaging systems. We are currently conducting cutting-edge molecular testing in metabolomics, proteomics, and single-cell sequencing, while leveraging artificial intelligence to analyze digital pathology slides.
In collaboration with BGI, we have conducted large-scale and in-depth multi-omics research, completed the whole genome sequencing analysis for approximately 14000 individuals, six susceptibility loci associated with non-small cell lung cancer have been replicated, and based on the analysis low-frequency and rare variations, 9 genes related to non-small cell lung cancer were identified. We have also profiled genome-wide genetic variation maps of a series of phenotypes. Additionally, we have carried out multi-omics research on lung cancer biomedical big data, identified multi-dimensional molecular profiles of lung cancer, constructed lung cancer biomarker clusters, and analyzed the molecular mechanism of lung cancer pathogenesis, so as to help the precise prevention and treatment of lung cancer. Using cutting-edge technologies such as metagenomics and metabolomics, a large database of precision nutrition organisms was constructed, and nutrition-related factors and pathways affecting diseases were mined. Through the fusion of cross-scale and multi-modal medical big data, we have conducted the multi-direction, multi-scale and multi-omics big data joint analysis, and applied the "dry-wet integration" approach to identify novel biomarkers related to the entire process of tumor to support the precise risk prediction.